For many laboratory researchers, the allure of automating data searches and seamlessly integrating them into their Electronic Laboratory Notebook to reclaim valuable time is no secret. But the question remains: How can they embark on this journey, and how do FAIR data principles play a pivotal role in this transformation? Read on to not only learn why but discover how.
## 1. What Are APIs and Why Are They Important?
Application Programming Interfaces, or APIs, are the backbone of digital interactions. Think of them as a set of rules that allow different software applications to communicate with each other. APIs are especially pivotal in the scientific world as they enable automation, enhance data integration, which in turn (and most importantly) saves valuable research time.
## 2. FAIR Data Principles and the Role of APIs
FAIR data principles aim to make scientific data Findable, Accessible, Interoperable, and Reusable (FAIR). APIs align perfectly with these goals by enabling seamless data transfer between platforms, automating routine tasks, and making data more accessible and usable.
## 3. Do I need to be a computer programming expert to use APIs in my laboratory?
No. While some automations or integrations may require the expertise of a developer, there are many tasks that, with some basic programming knowledge, can be built yourself. Learning a simple coding language doesn’t have to be time consuming or difficult.
## 4. Why Learn Python? A Simple Example with E-utilities to Find a Gene
Learning a simple coding language like Python can open doors to automating tasks and handling data more efficiently. For example, let's use Python and E-utilities to search for the gene "BRCA1" in the NCBI Gene database:
import requests
import json
# Define the API endpoint and query parameters
url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi"
params = {
"db": "gene",
"term": "BRCA1[Gene Name] AND Homo sapiens[Organism]",
"retmode": "json"
}
# Make the API request and parse the response
response = requests.get(url, params=params)
data = json.loads(response.text)
gene_ids = data['esearchresult']['idlist']
print(f"Gene IDs found for BRCA1 in Homo sapiens: {gene_ids}")
## 5. Using Labguru API to Create a Gene Entry
Labguru's API enables you to add new entries, like gene information, directly to your Electronic Lab Notebook (ELN). Here's how to create a new gene entry for "BRCA1":
# Authenticate to get your Labguru token
BASE_URL = 'https://your_labguru_domain'
auth_url = f"{BASE_URL}/api/v1/sessions.json"
credentials = {'login': 'your_email', 'password': 'your_password'}
auth_response = requests.post(auth_url, json=credentials)
auth_data = json.loads(auth_response.text)
auth_token = auth_data['token']
# Create a new gene entry
gene_data = {
'item': {
'name': 'BRCA1',
'sequence': 'ATCG...',
'description': 'Breast Cancer Gene'
},
'token': auth_token
}
headers = {'Authorization': f"Token token={auth_token}"}
gene_url = f"{BASE_URL}/api/v1/genes.json"
gene_response = requests.post(gene_url, headers=headers, json=gene_data)## 6. Combining E-utilities and Labguru APIs
Why not use both to make your research even more efficient? For example, you can search for "BRCA1" using E-utilities and use it to create an enriched entity in your Labguru ELN:
# First, fetch Gene IDs using E-utilities as before
# ...
# Then, use the Labguru API again to create the gene entry, now with the extra
information of the organism and the entrez id we got from E-utilities
gene_data['item']['entrez_id'] = gene_ids[0]
gene_data['item']['organism_name'] = "Homo sapiens"
gene_response = requests.post(gene_url, headers=headers, json=gene_data)
By integrating these two APIs, you're making your gene research more comprehensive and streamlined, fulfilling the FAIR data principles of being Findable, Accessible, Interoperable, and Reusable.
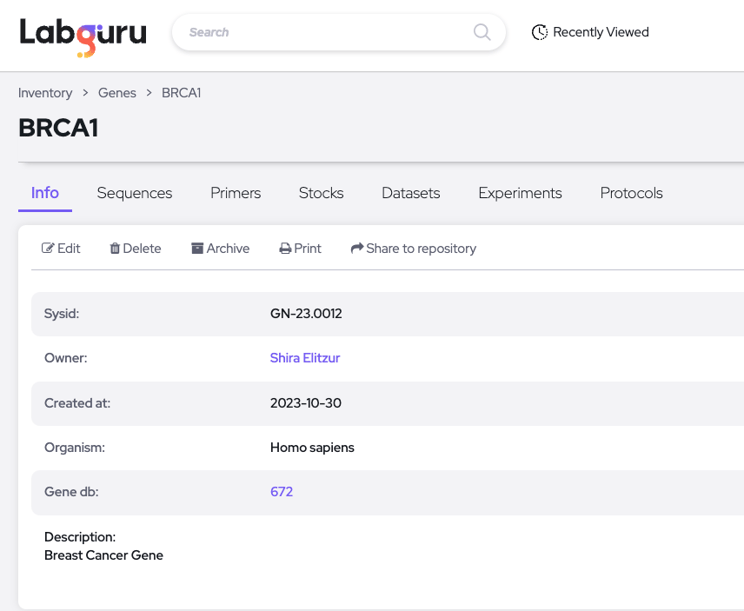
The actual new entity in your Labguru workspace:
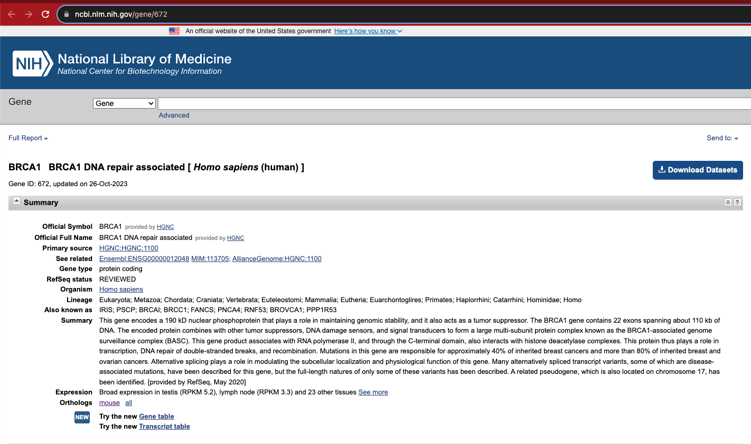
And it is easily linked to the corresponding NCBI entry:
APIs offer a pathway to not just automated but also FAIR-compliant research. With a little bit of Python and an understanding of how APIs like E-utilities and Labguru's can be combined, you can significantly enhance your research efficiency and data integrity.
Happy researching!
.png?height=380&name=fair%20data%20blog%20header%20(2).png)
%20(4).png)