Labguru ELN is very flexible, and self-explanatory which is a huge advantage for us. It allows each PI group to work on its own structure yet cooperate with other PI groups in the division. Labguru really stimulated the integration of our division and improved the collaboration and communication between our various groups.
Radboud University Medical Centre (Radboudumc) is a leading academic centre for medical science, education and health care, connecting research, education and patient care. located in Nijmegen, the Netherlands. Radboudumc mission is to have a significant impact on health care, aiming to be pioneers in shaping the health care of the future, in a person-centred and innovative way. Radboudumc, in total, has nearly 10,000 employees and 3,500 students. Roughly 20% of this group works in one of the research departments, varying from clinical research to lab-based research. Radboudumc currently has approximately 800 users working in Labguru, which is, approximately, over half of all employees working in their research labs.

We interviewed Remco Makkinje, BaS, a Coordinating Technician in the Human Genetics department. Remco is responsible for everything related to ICT and Data management in the Genome Research division. A very diverse job, being first line support for ICT, managing the lab information management system (LIMS) and related tools and being the workspace owner of Labguru. In addition, he does a bit of hands-on work in the lab.
Tell us about what the labs within Genome Research do? What kind of technics do you use in your kind of research?
Remco: The Genome Research division of the Department of Human Genetics consists of an international team of research technicians, geneticists, molecular biologists, bioinformaticians, statisticians operating at the frontiers of genetics in both research and teaching. The division is organized into 19 research groups led by Principal Investigators (PIs), who operate independently but collaboratively in performing research in the areas of neurobiology, sensory disorders, tumour genetics, and translational genomics. These PIs also play an important role as teachers and supervisors in the new curricula for Medicine and Biomedical Sciences, supported by a professional educational team.
The general aim of our division is to gain a better understanding of the mechanisms underlying human disease, with the goals of improving diagnosis, determining the right treatment for the right person, and developing novel therapeutic approaches.
Our research groups are structured into three main categories:
- Research groups primarily interested in biological mechanisms underlying specific disorders and how those can be used for treatment development. We aim to cover a broad range of diseases and disorders along the entire spectrum of genetically influenced morbidity in humans.
- Groups focused on developing novel methods for research and data analysis. In terms of method development, our main interest is in state-of-the-art genome research, with an important focus being the generation and use of Big Data through efficient and effective genomics, bioinformatics, and statistics pipelines.
- Groups developing and investigating cell and animal models. For the model systems, our division has made the choice to concentrate its efforts on the development of functional assays to investigate the biological effects of genetic variation observed in patients based on induced pluripotent stem cells/primary cells and small animal models, prioritizing fruit flies and zebrafish.
We use a lot of different techniques, starting from basic molecular techniques as PCR and gel electrophoresis and western blotting. But this extends to GWAS and Next Generation Sequencing (NGS) techniques as Exome or even Whole Genome Sequencing. For our functional work, we use a whole range of different protein interaction techniques, like Tandem affinity purification, proximity labelling and yeast-two-hybrid.
How did you start introducing and implementing Labguru within The Genome Research Department?
Remco: We were already looking for a replacement for our paper lab notebook for a while. At some point, all of the university hospitals in the Netherlands started a joint effort to test several ELN suppliers. Our hospital decided to test Labguru and our department was one of the departments where this was tested with a small number of users. We were satisfied with the very positive results, and we bought a hospital-wide license for Labguru.
We established a well-balanced implementation team consisting of several users from different PI-groups. We wanted a very flexible system which could be used to support all the different aspects of our research. With the full cooperation of the implementation team, it took us around 6 months to implement Labguru across the division.
What were the key challenges your department was facing?
Remco: With over 20 different PI-groups and around 125 active users, it was a real challenge to find a structured, as well as a flexible system, for our ELN. As every group has its own lab notebook working methods, we needed some flexibility. Together with the needed flexibility we also wanted a structured system with predefined rules to help us keep the data structured and organized.
Furthermore, finding the right balance between security and collaboration was a challenge. We didn’t want to give access to all the lab data to everyone. We only wanted data to be visible to people belonging to the same PI-group, not to the whole division. But then again, we had, of course, a lot of cooperation between different PI-groups, so that data needed to be accessible for people from multiple PI-groups. In addition, we also had general information which you want to share with everyone. Labguru allows us to create groups and define privilege-based access to projects.
What was your overall Goal when you started using Labguru?
Remco: Basically, we just wanted to get rid of our paper lab notebook. Genetics is a fast-moving field, with new bio-medical techniques such as next-generation genome sequencing that create vast amounts of data and require big data analysis capabilities. But somehow, in this fast-changing pace, we kept to our old ways concerning logging our lab work: printing out spreadsheets, gel pictures etc. and pasting them in our paper lab notebook. Finding out how older experiments were run or what primers were used in a specific experiment was a real hassle. We were interested in moving to an ELN and informatics system that will allow us to adapt our research to an era where research is done many times on the laptop and not on the workbench. We wanted an efficient and cost-effective system to keep track of our experiments.
While implementing, we also saw we could make a great improvement by replacing our current ordering database. Every PI group was using a separate system for ordering and this meant that we had to merge our ways of ordering and all our consumables we use in the lab.
How did Labguru help you?
Remco: The ELN section was a good match for us, as we like how it’s set-up. Labguru ELN is very flexible, and self-explanatory which is a huge advantage for us. It allows each PI group to work on its own structure yet cooperate with other PI groups in the division. Some groups use a lot of different projects, but others have just one or two projects and a lot of folders. The same goes for the experiment section, people can just keep track of the different experiments in the way they like. As we have very different techniques and different research topics flexibility is key.
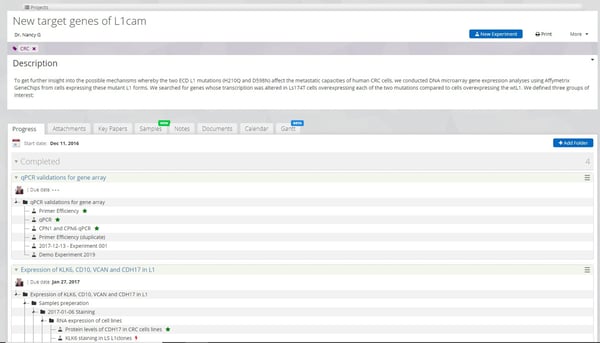
Labguru ELN projects management
The consumables inventory and shopping list were very helpful in order to replace our Access database ordering systems. It was easy to export the list of products and import them into Labguru using the import functionality. The management and ordering of products were comparable to how we did it in the previous system, but some out-of-the-box functionality was different. Luckily, it’s easy to add custom fields, so this way it’s easy to make this suitable for our needs.
How do you handle such a large number of users? How do you handle collaboration and communication? and how did Labguru helped?
Remco: Working in a large workspace means you need some form of structure. For this we set up some rules, we try to keep them limited in order to make it as open as possible. We have a unique 3-letter-code for instance for each PI-group, and the name of the title of every experiment, folder and project will start with this three-letter code. Each project is of course secured by making it visible only for the users related to the project. To check whether people stick to the rules we have ‘Superusers’. Each PI-group has a Superuser who oversees administration and provides instructions to new users. Every month we have a meeting with several Superusers to discuss improvements or issues regarding Labguru. This system works very well this way. We are glad we did it this way and not creating a workspace for each PI group, as this would have limited the collaboration between groups which we do a lot.
How do you deal with patients’ data organization and privacy and how does Labguru help you with that?
Remco: All our patients’ data gets pseudonymized when materials are coming in the lab. Personal identifiable information is not processed into Labguru, same as before when we used the paper notebook. We are also very sensitive about the other, non-personally identifiable, patient data we record in our lab notebook. We need to be very careful about where to store this. Therefore, together with Labguru, we set up a private encrypted cloud environment where all the Labguru data of the hospital is stored. In this way we sure all our patients’ data is stored in the safest way.
What Labguru features do you like most and find critical for your work?
Remco: The core functionality, the ELN, which was the first and foremost reason to implement Labguru. I like the way that you just start with one blank page and every user has its own way of putting their workflow into Labguru. It makes it very flexible and applicable to every type of experiment or study. I also like the user-friendliness and how intuitive the ELN is. New members only need a short introduction and the just learn by doing.
In addition, I think the shopping list and ordering system in Labguru is also something which is very useful. It’s an easy way to keep track of all the consumables and other goods. For instance, if you see something is low in stock in the lab it’s easy to find the product and ordering it. This is how everything should be, it’s as easy as buying something on Amazon. These kinds of things are key to keep the lab running in a successful way.
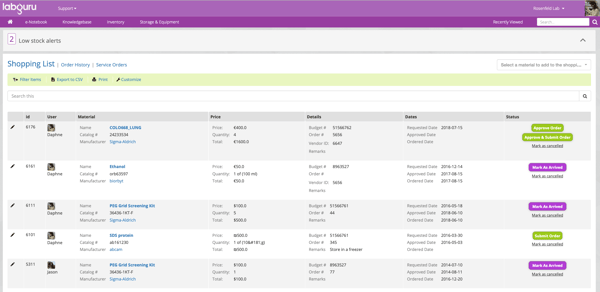
Labguru Shopping List feature
Can you describe the ROI you see from Labguru?
Remco: The biggest improvement was digitalizing the lab notebook; everything is now more structured. Things can be re-structured if necessary and information can now be retrieved more easily with Labguru’s great search functions. Moreover, Labguru really stimulated the integration of our division and improved the collaboration and communication between our various groups. For example, with Labguru, ordering, managing equipment, antibodies, etc., is much easier and cost effective as it is done on a division-wide level rather than separately for each group. Labguru allows us to get a better insight into what is done in each group.
To schedule a demo and learn more about Labguru – Contact Us